
| Microexon ID | Ps_NC_039358.1:228293342-228293345:- |
| Species | Papaver somniferum | Coordinates | NC_039358.1:228293342..228293345 |
| Microexon Cluster ID | MEP04 |
| Size | 4 |
| Phase | 0 |
| Pfam Domain Motif | Ribonuclease_T2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 51,4,53 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RGYWMWYCATCAACHTGCYWYRGTGGAAARGGAYYATTYTGGGCTCATGAGTGGGARAARCATGGRACWTGYTCYTCYCCWGTWRTTCRWGATGAATAYARYTAYTTY |
| Logo of Microexon-tag DNA Seq | 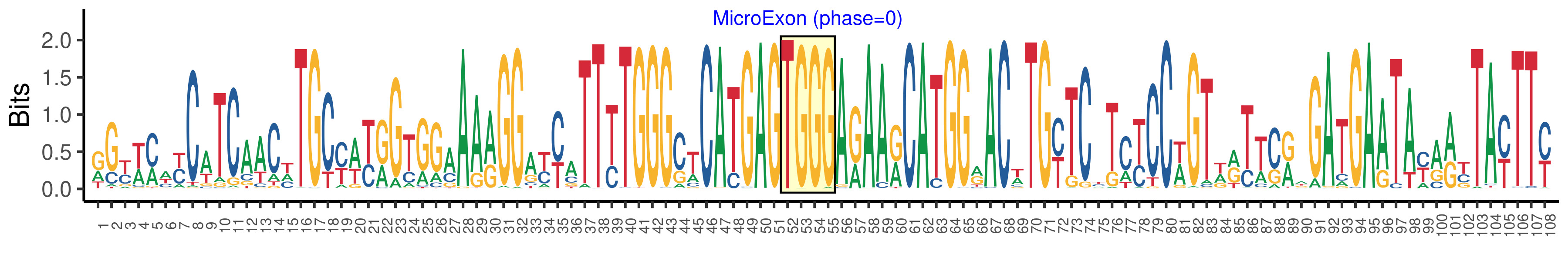 |
| Alignment of exons |  |
| Microexon DNA seq | TGGG |
| Microexon Amino Acid seq | WE |
| Microexon-tag DNA Seq | GGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCTGGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTC |
| Microexon-tag Amino Acid Seq | GKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYF |
| Microexon-tag spanning region | 228293104-228293529 |
| Microexon-tag prediction score | 0.9613 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026566295.1x |
| Reference Transcript ID | XM_026566295.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026566295.1 |
| Protein ID | XP_026422080.1 |
| Gene ID | LOC113318165 |
| Gene Name | NA |
| Pfam domain motif | Ribonuclease_T2 |
| Motif E-value | 1.3e-49 |
| Motif start | 40 |
| Motif end | 227 |
| Protein seq | >XP_026422080.1 MAFPKTLFVSVLLIVVSSCLFVEIDGRSRSRSLLSEQREFDYFALSLQWPGTICQHTRHCCAGNGCCRSDNGPTEFTIHG LWPDYNDGTWPACCTETRFDEKLISSLRKDLDKYWPSLSCGKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYFTTTL SVYFKYNVTAILAAEGYVPSNSEKYPLGGIVSAVQNAFGATPQVVCSHGAIEELRLCFYKDFKPRDCIMNEKHRSCPKYV SMPERRPMGHESSLPWSLDSEVFSS* |
| CDS seq | >XM_026566295.1 ATGGCTTTCCCTAAAACTCTTTTTGTCTCTGTTCTCTTGATTGTGGTGAGTTCTTGTTTGTTTGTCGAAATTGATGGTAG AAGTAGAAGCAGAAGTTTATTGAGTGAACAAAGAGAGTTTGATTACTTCGCTTTATCTCTTCAATGGCCTGGTACTATTT GTCAACATACTAGACATTGCTGTGCTGGTAATGGTTGTTGCAGAAGTGATAATGGACCAACGGAGTTCACAATCCATGGA CTCTGGCCCGACTACAATGATGGAACCTGGCCAGCATGTTGCACCGAAACTAGATTTGATGAGAAATTGATCTCATCATT GCGCAAAGATCTTGACAAGTATTGGCCGTCCTTAAGCTGTGGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCT GGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTCACAACGACTCTC AGTGTTTACTTCAAGTATAATGTCACGGCAATTTTGGCTGCCGAAGGATATGTACCCTCAAATTCTGAGAAATATCCCTT GGGAGGCATTGTTTCAGCCGTTCAAAATGCTTTTGGAGCAACCCCCCAGGTGGTTTGCTCACATGGTGCCATAGAGGAGC TTCGCTTATGCTTTTACAAAGATTTCAAGCCTCGCGACTGTATAATGAATGAAAAGCACAGGTCATGCCCGAAATACGTG AGCATGCCAGAACGCAGGCCGATGGGACATGAGTCTTCACTTCCATGGTCACTTGATAGTGAAGTATTCTCATCCTGA |
| Microexon DNA seq | TGGG |
| Microexon Amino Acid seq | WE |
| Microexon-tag DNA Seq | GGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCTGGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTC |
| Microexon-tag Amino Acid seq | GKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYF |
| Transcript ID | Ps.7202.2 |
| Gene ID | Ps.7202 |
| Gene Name | NA |
| Pfam domain motif | Ribonuclease_T2 |
| Motif E-value | 6.1e-39 |
| Motif start | 12 |
| Motif end | 165 |
| Protein seq | >Ps.7202.2 MVVAEVIMDQRSSQSVNGLWPDYNDGTWPACCTETRFDEKLISSLRKDLDKYWPSLSCGKPSTCSGGRGLFWAHEWEKHG TCSSPVVHDEYQYFTTTLSVYFKYNVTAILAAEGYVPSNSEKYPLGGIVSAVQNAFGATPQVVCSHGAIEELRLCFYKDF KPRDCIMNEKHRSCPKYVSMPERRPMGKFSRFIISSLLLGNYVNPYLGFYSVQGKSCLFISCINKGSYLMISKPSLLCRC GYIIRCYFVEHDTSRDHI* |
| CDS seq | >Ps.7202.2 ATGGTTGTTGCAGAAGTGATAATGGACCAACGGAGTTCACAATCCGTAAATGGACTCTGGCCCGACTACAATGATGGAAC CTGGCCAGCATGTTGCACCGAAACTAGATTTGATGAGAAATTGATCTCATCATTGCGCAAAGATCTTGACAAGTATTGGC CGTCCTTAAGCTGTGGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCTGGGCCCATGAGTGGGAAAAGCATGGC ACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTCACAACGACTCTCAGTGTTTACTTCAAGTATAATGTCAC GGCAATTTTGGCTGCCGAAGGATATGTACCCTCAAATTCTGAGAAATATCCCTTGGGAGGCATTGTTTCAGCCGTTCAAA ATGCTTTTGGAGCAACCCCCCAGGTGGTTTGCTCACATGGTGCCATAGAGGAGCTTCGCTTATGCTTTTACAAAGATTTC AAGCCTCGCGACTGTATAATGAATGAAAAGCACAGGTCATGCCCGAAATACGTGAGCATGCCAGAACGCAGGCCGATGGG TAAGTTTTCTAGATTCATCATTTCCTCTTTATTATTGGGTAATTACGTAAACCCCTATTTAGGTTTTTACAGTGTTCAAG GGAAAAGTTGCCTTTTCATTTCTTGCATTAATAAGGGTAGTTATTTGATGATTAGTAAACCTTCATTATTGTGCCGATGT GGCTACATAATACGGTGTTACTTTGTAGAACACGATACAAGTAGAGACCATATATGA |