
| Microexon ID | Sm_GL377586:1110547-1110549:- |
| Species | Selaginella moellendorffii | Coordinates | GL377586:1110547..1110549 |
| Microexon Cluster ID | MEP03 |
| Size | 3 |
| Phase | 0 |
| Pfam Domain Motif | CHORD |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 54,3,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCYAYSTTCACCGARGAYGAYAAYCCYGAMGRYTCCTGYMVWTAYCAYGMYTCKGGACCTWTATTTCATGATGGWATGAAAGARTGGAGYTGYTGCAARMAAAGAAGY |
| Logo of Microexon-tag DNA Seq | 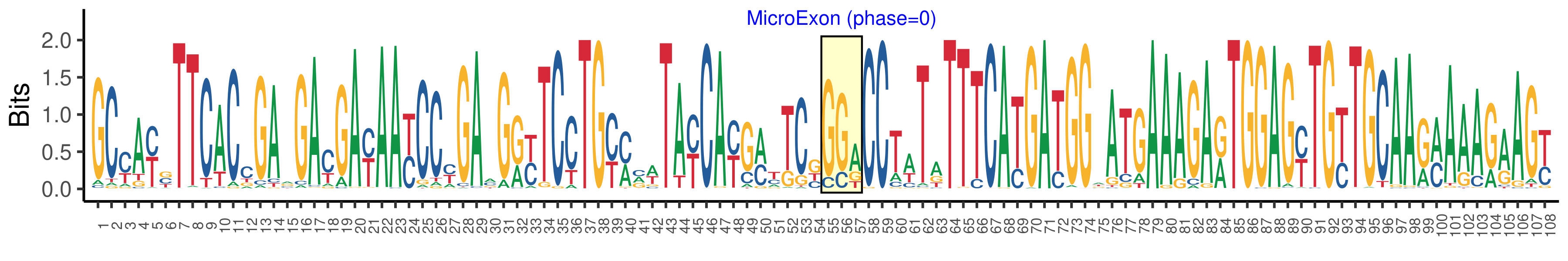 |
| Alignment of exons | 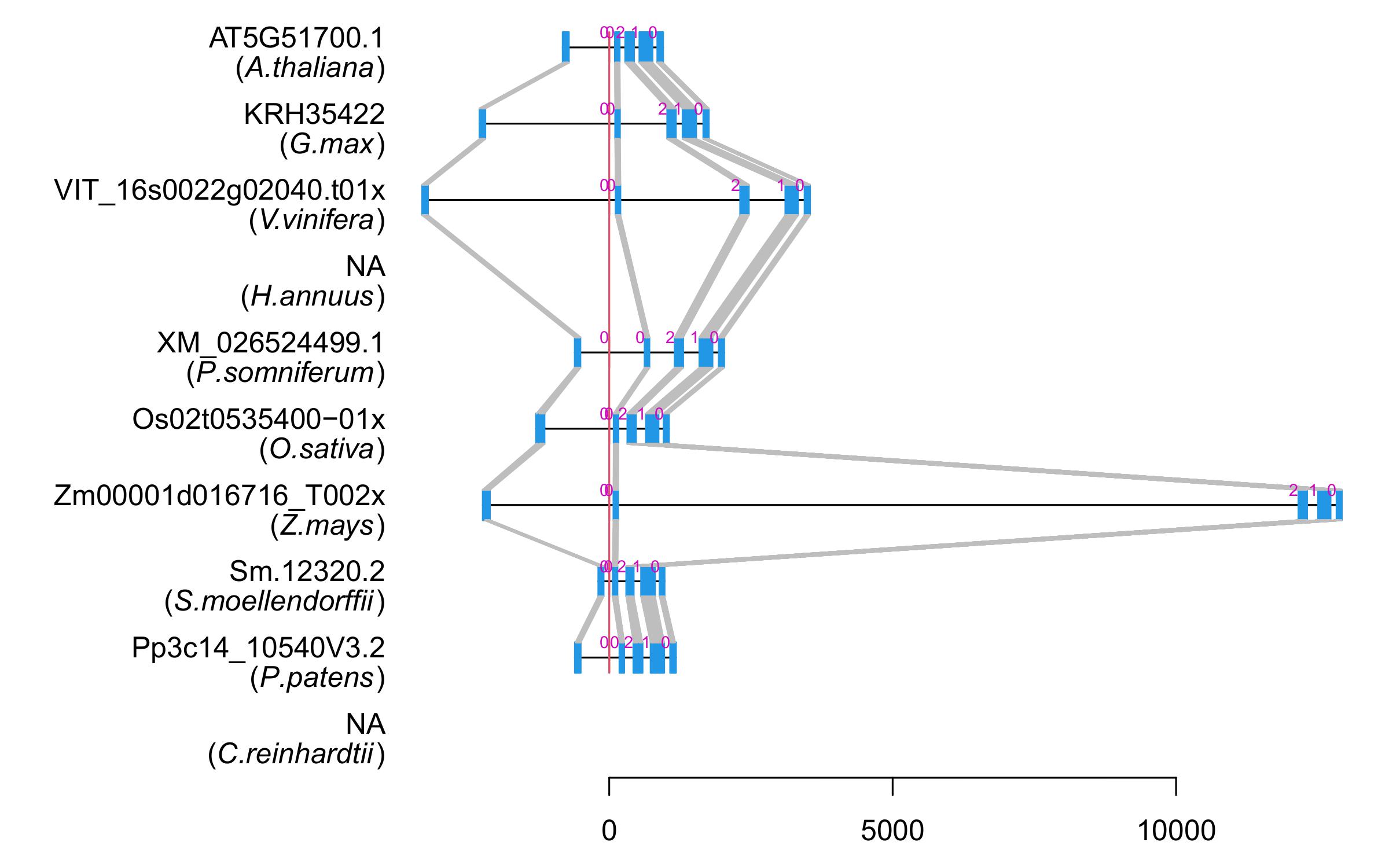 |
Sm_GL377586:1110547-1110549:- does not have available information here.
Sm_GL377586:1110547-1110549:- does not have available information here.
| Microexon DNA seq | CCG |
| Microexon Amino Acid seq | P |
| Microexon-tag DNA Seq | CTCTTCTCTCCCGATCAAAACGACGATGAGAAGGCTTGCTGCTACCATCCCGGCCCGCCCCTGTTTCACGACGGTGGGAAACAATGGAGTTGCTGCAAACAGCAGAGT |
| Microexon-tag Amino Acid seq | LFSPDQNDDEKACCYHPGPPLFHDGGKQWSCCKQQS |
| Transcript ID | Sm.12320.2 |
| Gene ID | Sm.12320 |
| Gene Name | NA |
| Pfam domain motif | CHORD |
| Motif E-value | 5e-24 |
| Motif start | 5 |
| Motif end | 66 |
| Protein seq | >Sm.12320.2 MGKVQCQRIGCDALFSPDQNDDEKACCYHPGPPLFHDGGKQWSCCKQQSHDFSLFLSIPGCTHGKHTSEKPKPAPKPSKT LPQGDASNTNCLRCKQGFYCSDHGSKISCGPAAAPSKDVEKPAAKEASQSSVQAAPAALPNVIDLDAKRTCRNKGCGKAF TERENHETACSYHPGPAVFHDRSRGWACCDVHVKEFDEFLEIPPCSKGWHSSQ* |
| CDS seq | >Sm.12320.2 ATGGGGAAGGTCCAATGCCAGCGGATTGGATGCGACGCGCTCTTCTCTCCCGATCAAAACGACGATGAGAAGGCTTGCTG CTACCATCCCGGCCCGCCCCTGTTTCACGACGGTGGGAAACAATGGAGTTGCTGCAAACAGCAGAGTCACGATTTTAGCT TGTTCTTAAGCATTCCAGGCTGTACGCACGGAAAGCACACATCAGAGAAACCAAAGCCAGCTCCCAAGCCAAGCAAAACT TTACCACAGGGAGACGCCAGCAACACCAATTGCTTGAGATGCAAACAAGGTTTCTACTGTTCAGATCACGGCTCAAAGAT ATCGTGTGGACCAGCGGCAGCCCCCAGCAAAGACGTGGAAAAACCAGCTGCGAAAGAAGCTTCGCAATCATCGGTGCAAG CAGCCCCGGCGGCCCTCCCCAACGTGATTGACTTGGATGCAAAGCGGACGTGCAGGAATAAAGGCTGCGGAAAGGCATTC ACCGAGCGAGAGAACCATGAAACTGCTTGCAGTTATCATCCGGGCCCAGCTGTTTTTCACGACAGATCGAGAGGATGGGC CTGCTGTGATGTTCATGTGAAGGAGTTTGACGAGTTCCTGGAGATCCCGCCGTGCTCGAAAGGCTGGCACAGCAGCCAAT AG |