
| Microexon ID | Ps_NC_039365.1:28146976-28146978:+ |
| Species | Papaver somniferum | Coordinates | NC_039365.1:28146976..28146978 |
| Microexon Cluster ID | MEP03 |
| Size | 3 |
| Phase | 0 |
| Pfam Domain Motif | CHORD |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 54,3,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCYAYSTTCACCGARGAYGAYAAYCCYGAMGRYTCCTGYMVWTAYCAYGMYTCKGGACCTWTATTTCATGATGGWATGAAAGARTGGAGYTGYTGCAARMAAAGAAGY |
| Logo of Microexon-tag DNA Seq | 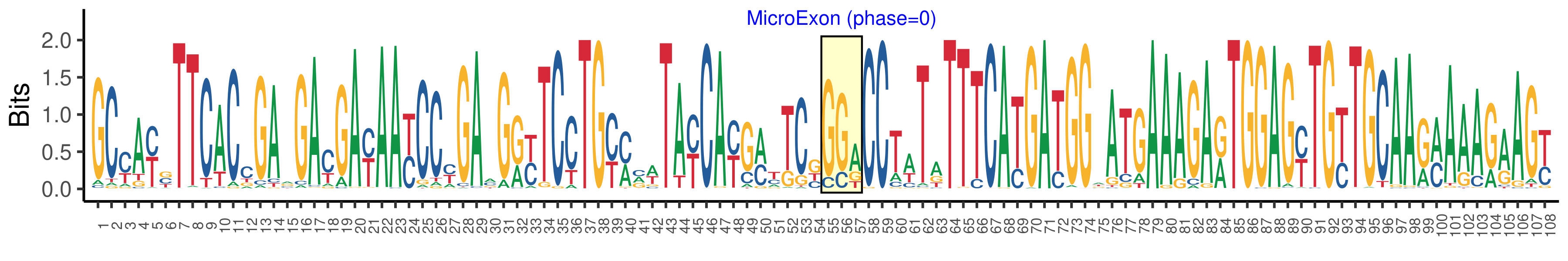 |
| Alignment of exons | 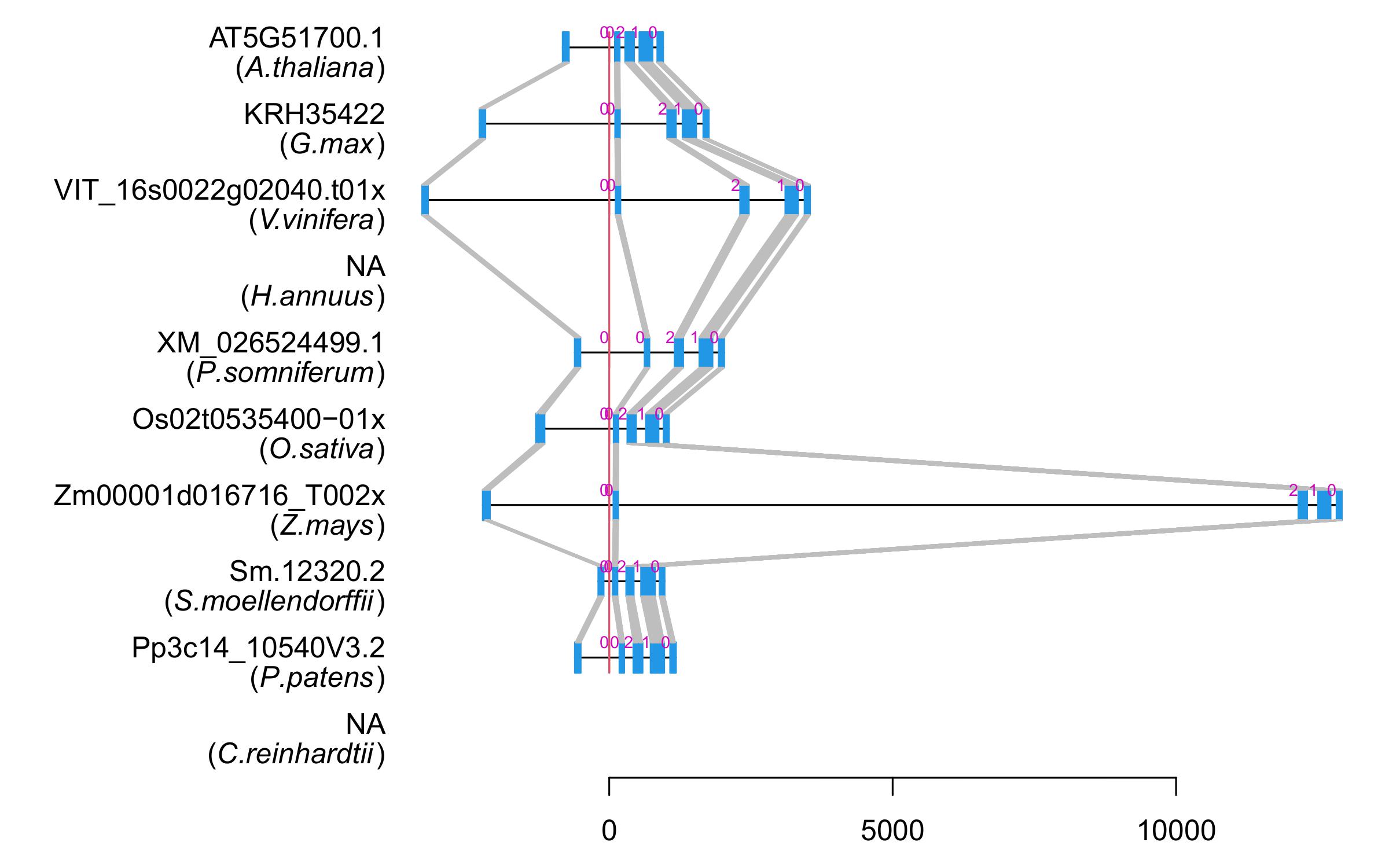 |
Ps_NC_039365.1:28146976-28146978:+ does not have available information here.
| Transcript ID | XM_026550202.1 |
| Protein ID | XP_026405987.1 |
| Gene ID | LOC113301419 |
| Gene Name | NA |
| Pfam domain motif | CHORD |
| Motif E-value | 7.1e-24 |
| Motif start | 8 |
| Motif end | 68 |
| Protein seq | >XP_026405987.1 MEKGSKVRCQRIGCDAMFTEDQNTEDSCTYHASGPIFHDGTKEWSCCKQRSHDFSLFLALPGCKTGKHTTEKPVIVKAAP KKPVPFVAPASTTSSKLDCSRCRQGFFCSEHGSQAKANTTMPTSPILPKCSTDVQEVTPPPAKKIININEPQTCRNKGCG RTFKEKDNSDDACSYHPGPAVFHDRMRGWKCCDVHVKEFDEFMEIPPCTKGWHSSDPIS* |
| CDS seq | >XM_026550202.1 ATGGAGAAGGGATCTAAGGTGAGATGTCAGAGAATAGGGTGTGATGCAATGTTCACTGAGGATCAAAATACTGAAGATTC TTGTACATATCATGCATCTGGGCCAATATTTCACGATGGTACGAAAGAGTGGAGCTGCTGCAAGCAAAGGAGTCACGATT TCAGCTTATTTTTAGCGCTTCCAGGATGCAAGACAGGTAAGCATACAACGGAAAAACCCGTGATTGTAAAAGCTGCACCG AAGAAGCCAGTTCCCTTTGTTGCTCCAGCAAGCACTACCTCATCAAAGCTGGATTGTTCAAGATGCCGACAGGGCTTCTT TTGCTCAGAACATGGTTCACAAGCTAAAGCAAACACAACTATGCCTACTTCTCCTATCCTCCCCAAGTGCAGTACCGACG TTCAGGAAGTCACACCCCCTCCTGCAAAAAAGATAATCAACATTAATGAACCTCAGACTTGCAGGAACAAAGGATGTGGC AGAACCTTCAAAGAAAAGGACAACAGTGATGATGCATGTAGTTACCACCCTGGTCCTGCCGTTTTCCACGACAGGATGAG AGGGTGGAAATGCTGCGATGTTCATGTGAAGGAGTTCGATGAGTTCATGGAAATACCTCCATGTACAAAGGGGTGGCATA GTTCAGATCCAATTTCTTAA |
| Microexon DNA seq | GGG |
| Microexon Amino Acid seq | G |
| Microexon-tag DNA Seq | GCAATGTTCACTGAGGATCAAAATACTGAAGATTCTTGTACATATCATGCATCTGGGCCAATATTTCACGATGGTACGAAAGAGTGGAGCTGCTGCAAGCAAAGGAGT |
| Microexon-tag Amino Acid seq | AMFTEDQNTEDSCTYHASGPIFHDGTKEWSCCKQRS |
| Transcript ID | XM_026550202.1 |
| Gene ID | Ps.48580 |
| Gene Name | NA |
| Pfam domain motif | CHORD |
| Motif E-value | 7.1e-24 |
| Motif start | 8 |
| Motif end | 68 |
| Protein seq | >XM_026550202.1 MEKGSKVRCQRIGCDAMFTEDQNTEDSCTYHASGPIFHDGTKEWSCCKQRSHDFSLFLALPGCKTGKHTTEKPVIVKAAP KKPVPFVAPASTTSSKLDCSRCRQGFFCSEHGSQAKANTTMPTSPILPKCSTDVQEVTPPPAKKIININEPQTCRNKGCG RTFKEKDNSDDACSYHPGPAVFHDRMRGWKCCDVHVKEFDEFMEIPPCTKGWHSSDPIS* |
| CDS seq | >XM_026550202.1 ATGGAGAAGGGATCTAAGGTGAGATGTCAGAGAATAGGGTGTGATGCAATGTTCACTGAGGATCAAAATACTGAAGATTC TTGTACATATCATGCATCTGGGCCAATATTTCACGATGGTACGAAAGAGTGGAGCTGCTGCAAGCAAAGGAGTCACGATT TCAGCTTATTTTTAGCGCTTCCAGGATGCAAGACAGGTAAGCATACAACGGAAAAACCCGTGATTGTAAAAGCTGCACCG AAGAAGCCAGTTCCCTTTGTTGCTCCAGCAAGCACTACCTCATCAAAGCTGGATTGTTCAAGATGCCGACAGGGCTTCTT TTGCTCAGAACATGGTTCACAAGCTAAAGCAAACACAACTATGCCTACTTCTCCTATCCTCCCCAAGTGCAGTACCGACG TTCAGGAAGTCACACCCCCTCCTGCAAAAAAGATAATCAACATTAATGAACCTCAGACTTGCAGGAACAAAGGATGTGGC AGAACCTTCAAAGAAAAGGACAACAGTGATGATGCATGTAGTTACCACCCTGGTCCTGCCGTTTTCCACGACAGGATGAG AGGGTGGAAATGCTGCGATGTTCATGTGAAGGAGTTCGATGAGTTCATGGAAATACCTCCATGTACAAAGGGGTGGCATA GTTCAGATCCAATTTCTTAA |