
| Microexon ID | Ps_NC_039366.1:124414579:- |
| Species | Papaver somniferum | Coordinates | NC_039366.1:124414579..124414579 |
| Microexon Cluster ID | MEP02 |
| Size | 1 |
| Phase | 1 |
| Pfam Domain Motif | Vps55 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 19,1,52 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | ATGGCGATGACMRTYGAAGTGCTTGCTGGRCTTGCTTTYATGTTYTCADCBAGYATYYTGYTRCARATMCTG |
| Logo of Microexon-tag DNA Seq | 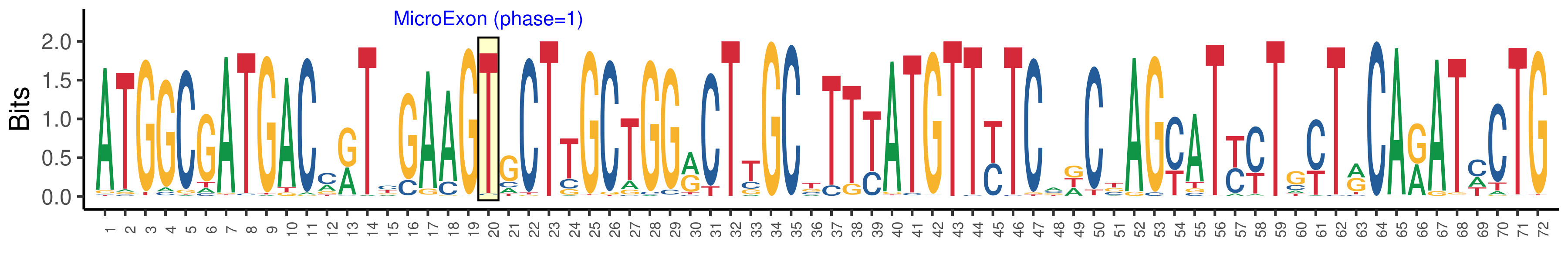 |
| Alignment of exons | 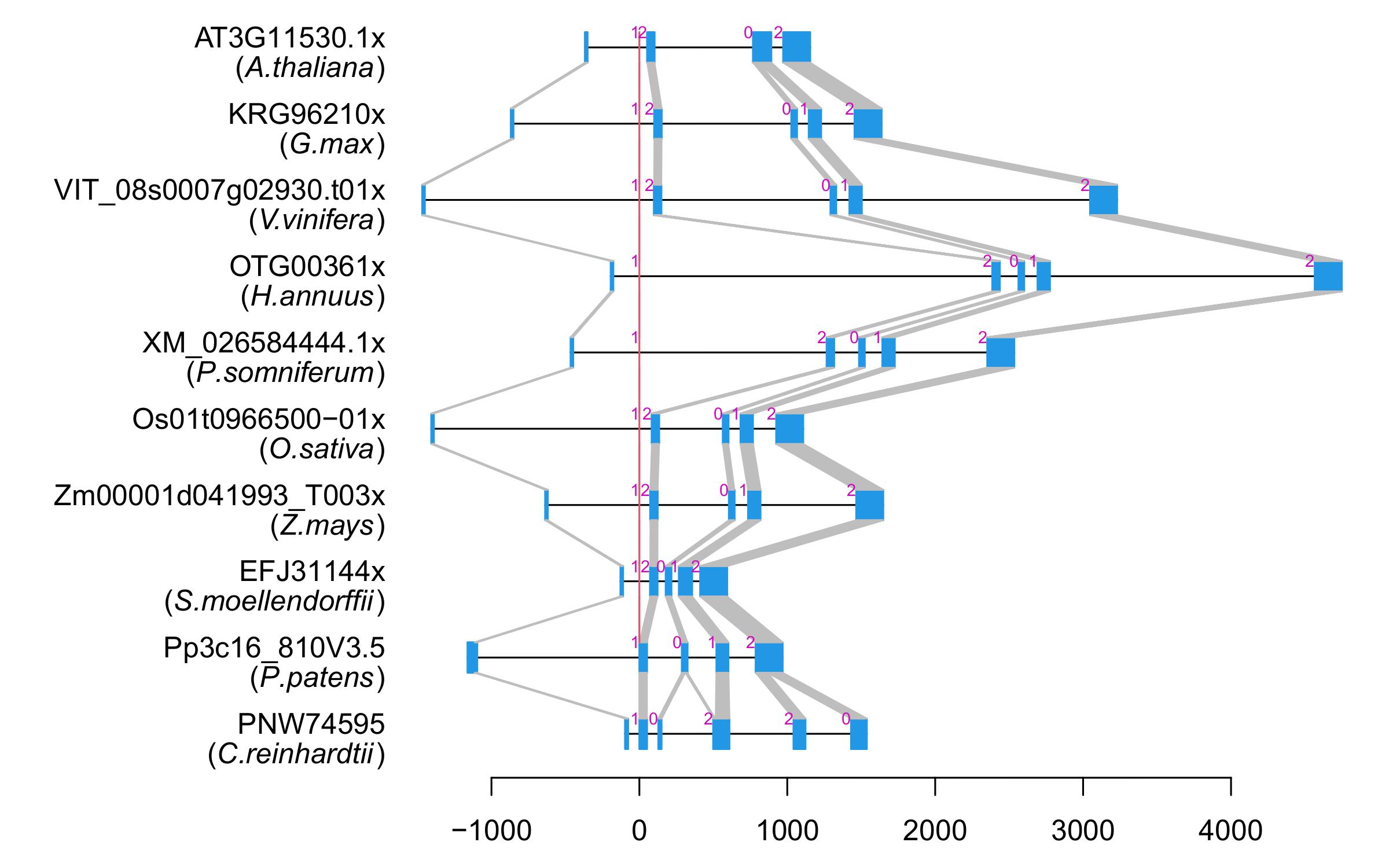 |
| Microexon DNA seq | T |
| Microexon Amino Acid seq | V |
| Microexon-tag DNA Seq | ATGGCTATGTCGGTCGAAGTGCTTGCAGGGCTGGCTTTCATGTTCTCAGCTAGCATTTTGTTGCAAATTCTG |
| Microexon-tag Amino Acid Seq | MAMSVEVLAGLAFMFSASILLQIL |
| Microexon-tag spanning region | 124413277-124415017 |
| Microexon-tag prediction score | 0.9604 |
| Overlapped with the annotated transcript (%) | NA |
| New Transcript ID | NA |
| Reference Transcript ID | NA |
| Gene ID | NA |
| Gene Name | NA |
Ps_NC_039366.1:124414579:- does not have available information here.
| Microexon DNA seq | T |
| Microexon Amino Acid seq | V |
| Microexon-tag DNA Seq | ATGGCTATGTCGGTCGAAGTGCTTGCAGGGCTGGCTTTCATGTTCTCAGCTAGCATTTTGTTGCAAATTCTG |
| Microexon-tag Amino Acid seq | MAMSVEVLAGLAFMFSASILLQIL |
| Transcript ID | Ps.57094.1 |
| Gene ID | Ps.57094 |
| Gene Name | NA |
| Pfam domain motif | Vps55 |
| Motif E-value | 4e-31 |
| Motif start | 9 |
| Motif end | 120 |
| Protein seq | >Ps.57094.1 MAMSVEVLAGLAFMFSASILLQILACAIYSNWWPMLAALMYVMVPMPCLFFGGGSTAFLTSRDGGGWMDAAKFLTGASAV GSIAIPAILRHAGLIETGAMFIEFTSFFILVCTIMCFHRASLEDEW* |
| CDS seq | >Ps.57094.1 ATGGCTATGTCGGTCGAAGTGCTTGCAGGGCTGGCTTTCATGTTCTCAGCTAGCATTTTGTTGCAAATTCTGGCCTGTGC AATTTACAGCAACTGGTGGCCGATGTTGGCAGCGCTCATGTACGTGATGGTACCTATGCCTTGTTTATTTTTTGGTGGTG GATCCACTGCATTCTTAACCAGCCGTGATGGTGGAGGCTGGATGGATGCTGCGAAGTTCTTGACAGGAGCATCCGCAGTA GGAAGCATCGCCATTCCTGCAATCCTTAGGCATGCTGGTCTGATCGAAACAGGAGCTATGTTCATTGAGTTCACTTCTTT CTTCATTCTTGTTTGCACTATCATGTGTTTTCATCGTGCTAGTCTTGAAGATGAATGGTGA |