![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | S12_GGCTAC_L008_R1_001.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 32776281 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 48 |
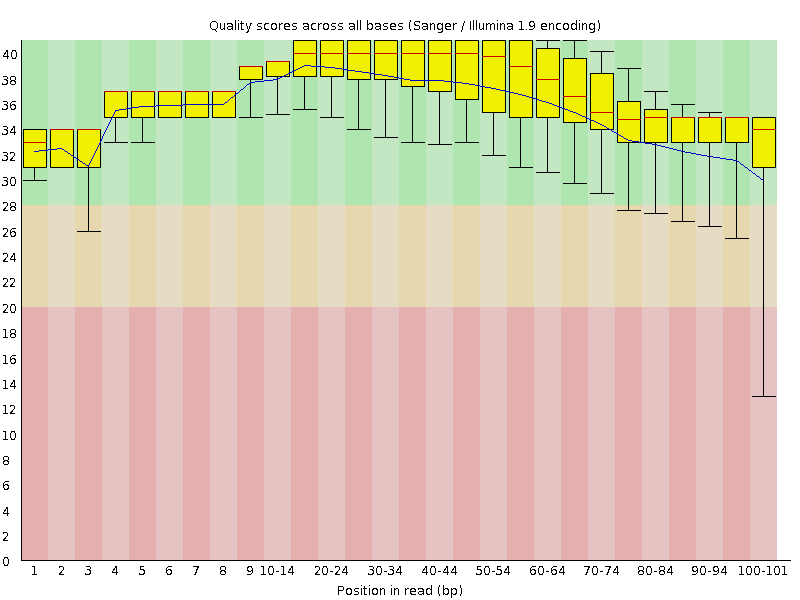
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
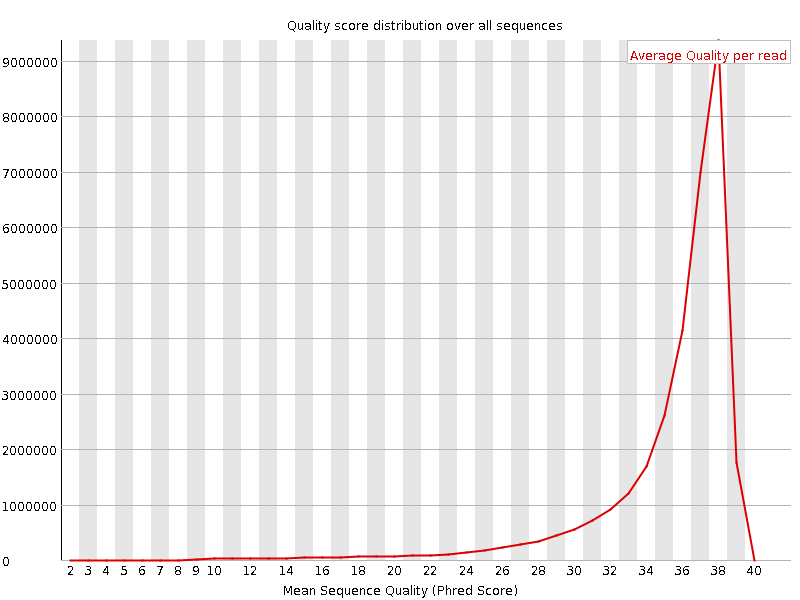
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
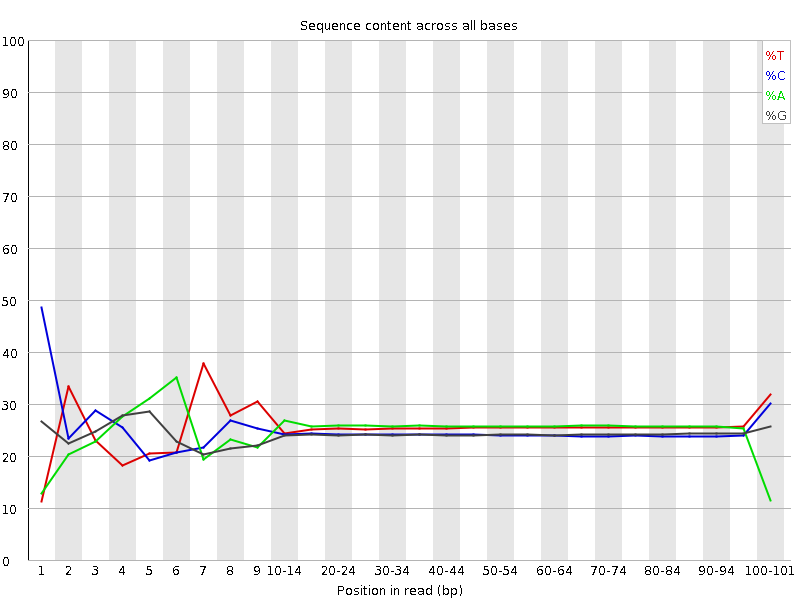
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
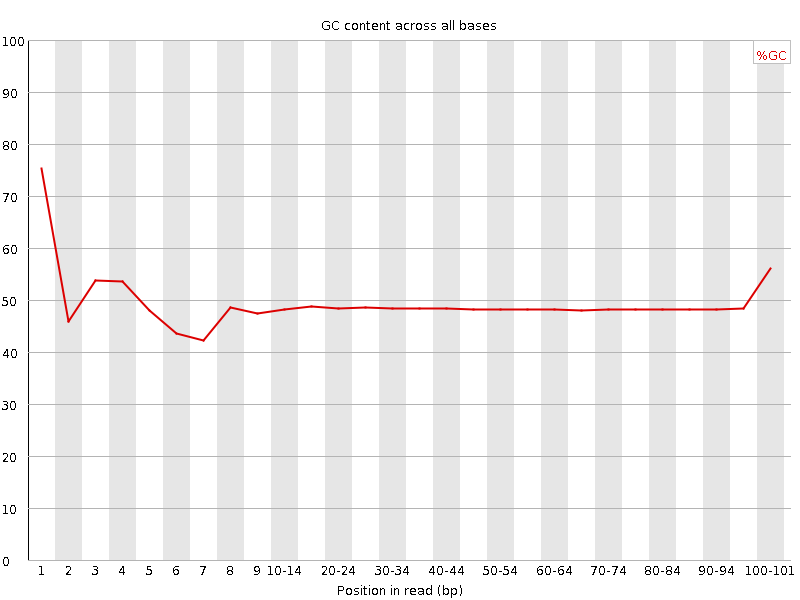
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[FAIL]](Icons/error.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
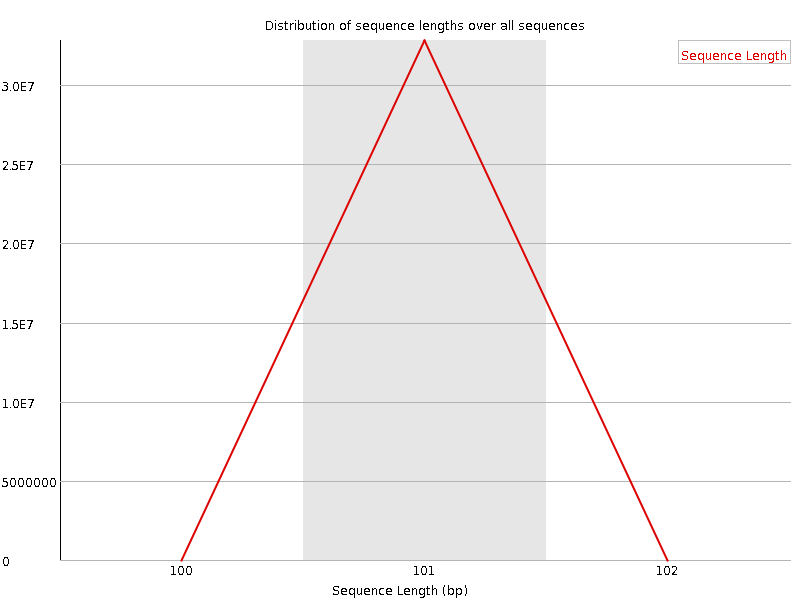
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGGCTACATCTCGTATGC | 47327 | 0.14439405129581356 | TruSeq Adapter, Index 11 (100% over 50bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
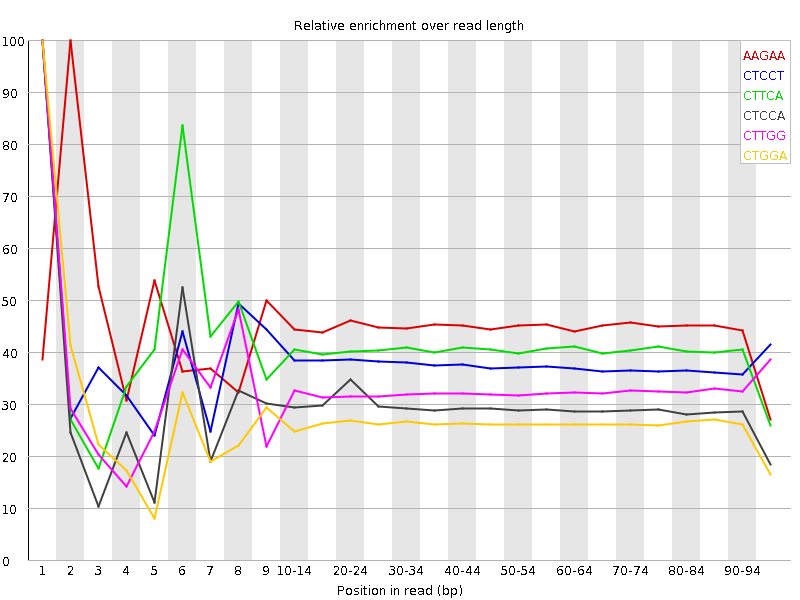
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAGAA | 8192495 | 2.5055523 | 5.5847106 | 2 |
| CTCCT | 6223875 | 2.0578396 | 5.4240894 | 1 |
| CTTCA | 6452060 | 2.0428054 | 5.000954 | 1 |
| CTCCA | 5956855 | 1.9708014 | 6.6691747 | 1 |
| CTTGG | 4993775 | 1.67587 | 5.1010623 | 1 |
| CTGGA | 4877380 | 1.6378464 | 6.1415763 | 1 |